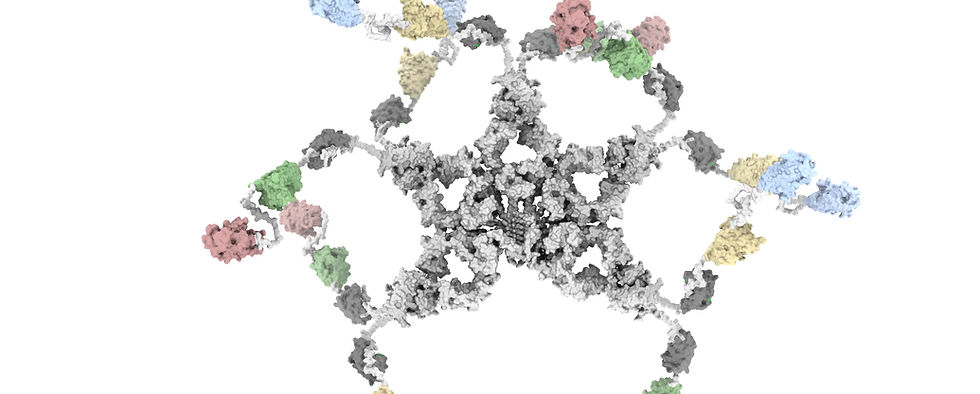
We are interested in advancing biomedical science by developing technologies that redefine the boundaries of discovery. We develop next-generation antibodies with unmatched precision to diagnose and treat infectious diseases, cancer, and autoimmune disorders. Leveraging cutting-edge tools in protein engineering and design, artificial intelligence, and proteomics, we aim to uncover novel biology and accelerate the drug discovery process. Through interdisciplinary collaboration, we bridge the gap between basic science and translational medicine, delivering innovative solutions to some of the most pressing challenges in human health. Our lab is dedicated to fostering a collaborative and inclusive environment where curiosity, creativity, and scientific rigor drive groundbreaking advancements.
Research Statement
11/27/2025: Congrats Yufei on the Science paper with Marty Taylor at Brown! Lovely work showing how mTORC2 recognizes its substrates.
11/25/2025: Huge congrats on Frank and Zhe's Cell Systems paper! A beautifully crafted and significant computational biology study that reveals near-universal immune escape by SARS-CoV-2 antibodies and the extraordinary challenges of mitigating viral escape with conventional antibody technologies.
Best wishes Frank on your PhD applications!
09/01: Celebrating our two amazing high school gals, Alba and Willa, for their lovely summer internship. STEM way to go!!!!!!
07/25/2025: Successful thesis defense by Zhe! Huge congrats to Dr. Sang on receiving the Doctor of Distinction, adding another remarkable accolade to the group!🎉🎉🎉
06/13/2025: Successful thesis defense by Jeff! Hats off to Dr. Kim on earning the Doctor of Distinction honor from Mount Sinai! Best wishes Jeff on your residency at Pitt! 🎉🎉🎉
10/23/2024: Our AMETA technology is now published at Cell. Congratulations Yufei, Zhe and Pham! What an incredible journey!!!